2 Quality Control
To begin work on producing a set of PGSs, the input data must first be carefully controlled. Throughout this project the UK Biobank will be the solitary set of data examined. The reason being is that the UK Biobank is the only large and easily accessed data-set that contains both genotypic and phenotypic information.
A brief description of the UK Biobank …
2.1 Previously Applied QC
In the process of sequencing the data the good people at the UK Biobank have already applied some quality control measures. To understand exactly what they did we first need to know how they even got the data to begin with. Of the nearly 500,000 people that were avaliable to be sequenced, batches of approximitley 4,600 people were created. The first 11 batches were sequenced on the UK BiLEVE Axiom array, the remaining 95 batches were sequenced with the UK Biobank Axiom Array. Both arrays were carefully created for this type of research, although we should be careful going forward as batch effects could change allele frequencies and the ultimate PGS distribution.
As the arrays were novel, some of the allele probes were not constructed well enough to clearly determine what allele a person was. A total of 35,014 unique markers were therefore removed from everyone right off the bat. Genotype calling by Affymetrix resulted in a data set of 489,212 individuals typed at 812,428 markers with which to carry out further QC. This QC was population structure aware, with different ancestry groups determined by comparing PCA loadings from UK Biobank individuals to a selection of 1000 Genomes Project individuals (The markers that went into this PCA were carefully QC’d). Afterwards, within population structure homogenous groups to each batch the following tests were applied:
- Batch Effects
- Plate Effects
- Departures from Hardy-Weinberg Equilibrium
- Sex Effects
- Array Effects
- Discordance Across Controls
If a marker failed the first 4 tests it was removed from the batch, and if a marker failed the last 2 tests then it was removed from the entire data-set. Each of these tests are hypothesis tests, with the cut-off chosen for removal set at p < 10^-12. Clearly this is pretty extreme so it is important to note there may be more QC that needs to be done.
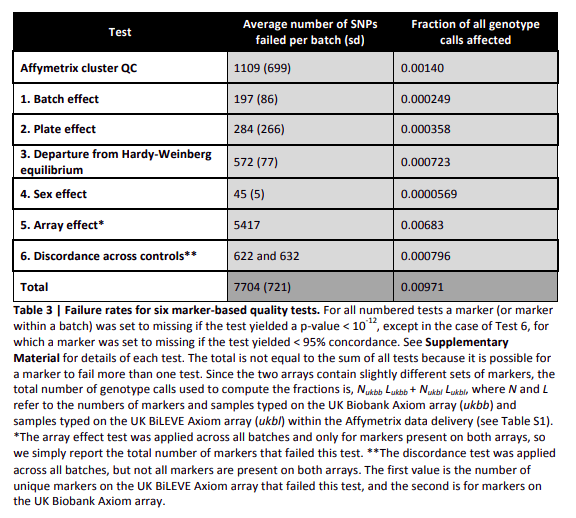
Figure 2.1: Failure rates
Following marker QC a pipeline of sample QC was applied. A good description of the pipeline is shown below. What is important to note is that the doccumentation seems to note that only individuals with really extreme missingness or hetrozygosity were removed from the dataset. Otherwise they were simply marked as being an outlier in their respective test. A more stringent threshold of the sample QC and marker QC was set for the dataset that was used within a principal component analysis. The output of this PCA was then used to determine a white, British ancestry subset. This was completed by comparing the samples who declared they were white and British and then removing outliers from the main cluster of this grouping based on PCA.
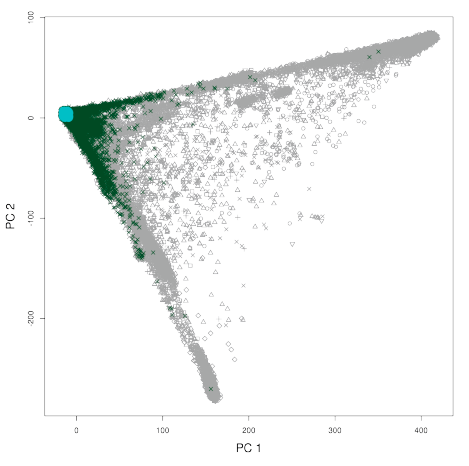
Figure 2.2: British PCA
Far more information on all of these steps can be found from the BioRxiv pdf: “Genome-wide genetic data on ~500,000 UK Biobank participants”. The big takeaways that I gleam are that the UK Biobank did think about genotyping seriously, the most abberant markers have been removed although some questionable markers likely remain, and that quite a few abberant samples remain although removing them should be easy since they have been marked. All of this gives a good starting point for this work.
2.2 Initial QC
To increase levels of QC, a file that lists all of the outliers was used to create a list of high quality anonymous IDS. The script that ran this entire process is displayed below.
In short, the process carried out involved removes outliers in hetrozygosity, putative sex chromosome aneupolidy, and excess relatvies all determined by the UK Biobank processes described above. Lastly, only individuals who were determined to be white with British ancestry were kept. This is a highly unfortunate cut-off, but nescessary in order to create a genetically homogenous individuals to be tested.
Now that we have a base level of genetic data QC, we can move onto QC of the summary statistics.